Spindles detection and analysis
Source:vignettes/Spindles_detection_and_analysis.Rmd
Spindles_detection_and_analysis.RmdSleep spindles are distinct bursts of brain activity that occur mostly during stage 2 of non-rapid eye movement (NREM) sleep. Characterized by their short duration, typically lasting between 0.5 to 2 seconds, they appear as a rapid series of waves on an electroencephalogram (EEG). These waves are typically in the 11 to 16 Hz frequency range De Gennaro and Ferrara (2003) . Sleep spindles are thought to play a crucial role in brain development, memory consolidation, and cognitive function. Their presence and characteristics can vary depending on age, cognitive factors, and certain neurological conditions, making them a significant focus in sleep research and neuroscience.
While EEG-based visual inspection remains the benchmark for identifying these spindles, this process is not only labor-intensive and expensive but also prone to variability and bias among different scorers. An algorithm for sleep spindle detection significantly streamline this process, enhancing both efficiency and consistency in spindle identification.
The A7 algorithm Lacourse et al. (2019) operates using a single EEG channel and is dependent on four objective key parameters: the absolute power of the sigma band, its relative power, and the correlation or covariance between the sigma band-passed signal and the original EEG signal.
Sleep data
EEG signal
First, download and read an single EEG signal from an example EDF Kemp et al. (1992) file. According to the standardized 10-20 system Niedermeyer and Lopes da Silva (2005), the C3-M2 derivation refers to the measurement of electrical brain activity recorded between the C3 electrode position on the left side of the head and the M2 electrode positioned on the right mastoid.
library(edfReader)
download.file(
url = "https://rsleep.org/data/15012016HD.edf",
destfile = "15012016HD.edf")
h <- readEdfHeader("15012016HD.edf")
s <- readEdfSignals(h, signals = "C3-M2")#> [1] TRUEScored events
Then, download and read the scored events containing the hypnogram data to epoch the signal and focus the analysis on N2 epochs.
download.file(
url = "https://rsleep.org/data/15012016HD.csv",
destfile = "15012016HD.csv")
events <- rsleep::read_events_noxturnal("15012016HD.csv")#> [1] TRUEThe hypnogram can be plotted using plot_hypnogram() function from rsleep.
plot_hypnogram(events)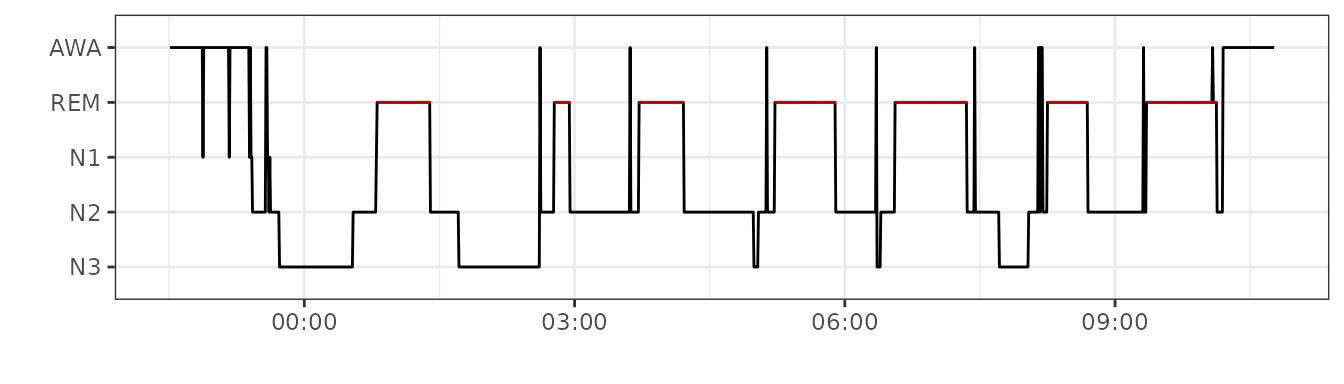
Spindle detection using A7
Select the 10th N2 epoch that we know contain spindles.
n2_epoch = events[events$event == "N2",][10,]Compute the epoch boundaries in the signal: subtract the epoch times from the signal start time, then multiply by the signal’s sample rate. Add one to the start result to adjust for R’s indexing system.
n2_epoch_index_start = as.numeric(
difftime(n2_epoch$begin, s$startTime, units = "secs")) * s$sRate + 1
n2_epoch_index_end = as.numeric(
difftime(n2_epoch$end, s$startTime, units = "secs")) * s$sRateGet the signal and multiply it as the A7 algorithm was designed for uVolts and the current EEG record is in Volts.
eeg = s$signal[n2_epoch_index_start:n2_epoch_index_end]
eeg = eeg * 1000000Run the A7 algorithm implemented in rsleep:
result = a7(
x = eeg,
s$sRate)
Display the 3 first detected spindles properties.
result$spindles[1:3,]Visualise the second detected spindle with ggplot2.
library(ggplot2)
data = data.frame(x=eeg,index=seq_along(eeg))
a = result$spindles$idxStart[2]
b = result$spindles$idxEnd[2]
data = data[(data$index <= (b+600)) & (data$index >= (a-600)), ]
ggplot(data, aes(x = index, y = x)) +
geom_line() +
geom_line(data = subset(data, index >= a & index <= b), aes(x = index, y = x), color = "red") +
labs(x = "Signal index", y = "C3-M2") +
theme_minimal()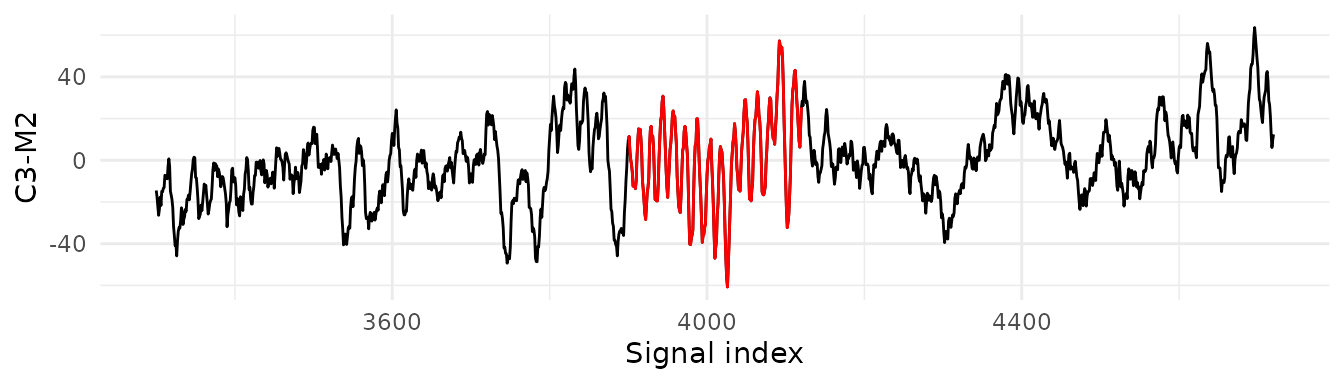
Overnight spindle detection
Joining the previous steps in a for loop along all N2 epochs, spindles are detected over the whole sleep record:
n2_epochs = events[events$event == "N2",]
epochs_results = list()
for(i in c(1:nrow(n2_epochs))){
n2_epoch_index_start = as.numeric(
difftime(n2_epochs$begin[i],s$startTime,units = "secs")) * s$sRate + 1
n2_epoch_index_end = as.numeric(
difftime(n2_epochs$end[i],s$startTime,units = "secs")) * s$sRate
eeg = s$signal[n2_epoch_index_start:n2_epoch_index_end]
eeg = eeg * 1000000
results = rsleep::a7(
x = eeg,
s$sRate)
epochs_results[[length(epochs_results)+1]] = results
}Detected spindles from all the epochs may be bound together using bind_rows() from dplyr.
spindles = dplyr::bind_rows(
lapply(c(1:length(epochs_results)), function(x){
epochs_results[[x]]$spindles$epoch = x
epochs_results[[x]]$spindles
}))Finally, plot the number of detected spindles by N2 epoch number using ggplot2:
library(ggplot2)
ggplot(spindles, aes(x = epoch)) +
geom_bar() +
xlab("N2 epoch number") +
ylab("Detected spindles")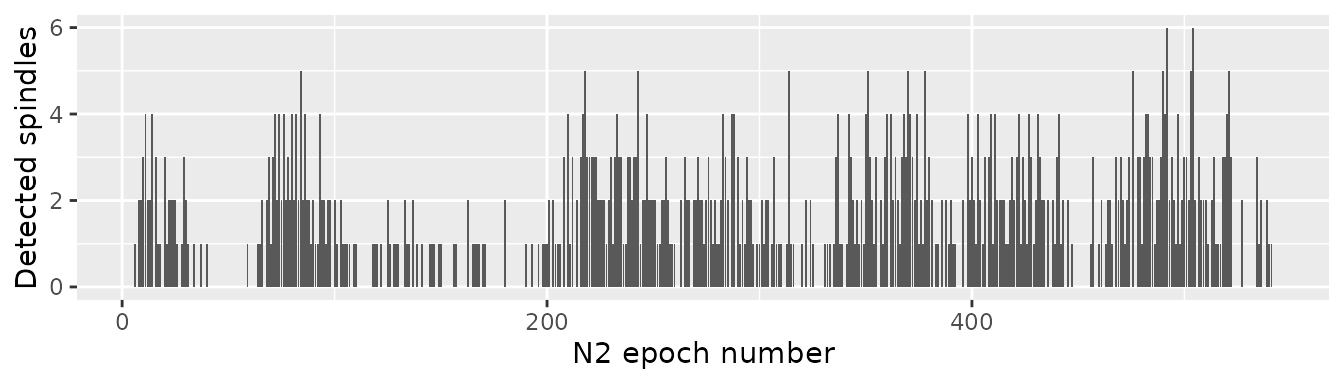
De Gennaro, Luigi, and Michele Ferrara. 2003. “Sleep Spindles: An Overview.” Sleep Medicine Reviews 7 (5): 423–40. https://doi.org/10.1053/smrv.2002.0252.
Kemp, Bob, Alpo Värri, Agostinho C. Rosa, Kim D. Nielsen, and John Gade. 1992. “A simple format for exchange of digitized polygraphic recordings.” Electroencephalography and Clinical Neurophysiology 82 (5): 391–93. https://doi.org/10.1016/0013-4694(92)90009-7.
Lacourse, Karine, Jacques Delfrate, Julien Beaudry, Paul Peppard, and Simon C. Warby. 2019. “A Sleep Spindle Detection Algorithm That Emulates Human Expert Spindle Scoring.” Journal of Neuroscience Methods 316 (March): 3–11. https://doi.org/10.1016/j.jneumeth.2018.08.014.
Niedermeyer, Ernst, and F. H. Lopes da Silva, eds. 2005. Electroencephalography: Basic Principles, Clinical Applications, and Related Fields. 5th ed. Philadelphia: Lippincott Williams & Wilkins.